partaker
Partaker
Partaker is a tool to analyze bacterial populations in microfluidic environments, via timelapse microscopy. With Partaker, you can generate plots such as:
Tutorials
Currently a WIP, we are creating a series of tutorials on how to use the functionality of this tool:
- Tool basics, loading an experiment, navigating, segmenting images
- Population Fluorescence dynamics analysis
Installation
The installation is quite straightforward as stated in the README.
Configuring VS Code for development
Set your launch.json to be something like this:
{
"version": "0.2.0",
"configurations": [
{
"name": "Python Debugger: nd2-analyzer",
"type": "debugpy",
"request": "launch",
"module": "nd2_analyzer",
"console": "integratedTerminal",
"cwd": "${workspaceFolder}/src",
"env": {
"PARTAKER_GPU": "1",
"UNET_WEIGHTS": "/Users/hiram/Documents/EVERYTHING/20-29 Research/22 OliveiraLab/22.12 ND2 analyzer/nd2-analyzer/src/checkpoints/delta_2_20_02_24_600eps"
},
"python": "${command:python.interpreterPath}"
}
]
}
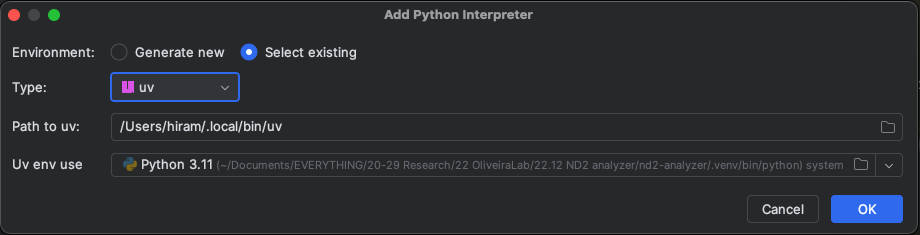
Configuring PyCharm for development
Quite straightforward as well. In project, select a Python Interpreter, and use the uv configuation and the venv you created for this project: